Multifractal Analysis for Evaluating the Representation of Clouds in Global Kilometer‐Scale Models
Geophysical Research Letters Wiley 51:20 (2024) e2024GL110124
Abstract:
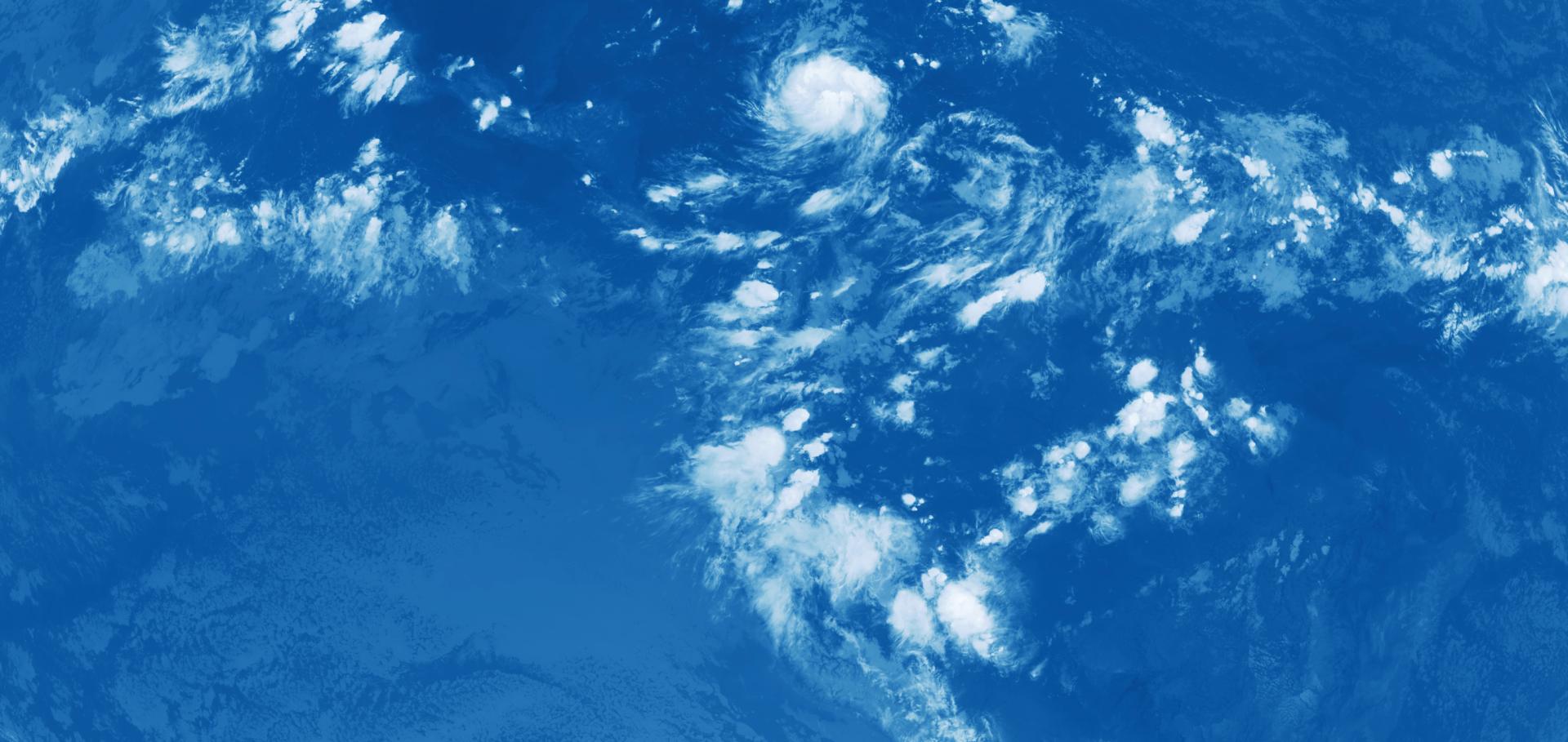
Clouds are one of the largest sources of uncertainty in climate predictions. Global km‐scale models need to simulate clouds and precipitation accurately to predict future climates. To isolate issues in their representation of clouds, models need to be thoroughly evaluated with observations. Here, we introduce multifractal analysis as a method for evaluating km‐scale simulations. We apply it to outgoing longwave radiation fields to investigate structural differences between observed and simulated anvil clouds. We compute fractal parameters which compactly characterize the scaling behavior of clouds and can be compared across simulations and observations. We use this method to evaluate the nextGEMS ICON simulations via comparison with observations from the geostationary satellite GOES‐16. We find that multifractal scaling exponents in the ICON model are significantly lower than in observations. We conclude that too much variability is contained in the small scales ( < 100 k m ) $(< 100\ \mathrm{k}\mathrm{m})$ leading to less organized convection and smaller, isolated anvils.Multifractal Analysis for Evaluating the Representation of Clouds in Global Kilometre-Scale Models
(2024)
Multifractal analysis for evaluating the representation of clouds in global km-scale models
Copernicus Publications (2024)
Multifractal Analysis for Evaluating the Representation of Clouds in Global Kilometre-Scale Models
(2024)
A Machine Learning Approach for Predicting Essentiality of Metabolic Genes
In: Braman, J.C. (eds) Synthetic Biology. Methods in Molecular Biology, vol 2760 (2024)
Abstract:
The identification of essential genes is a key challenge in systems and synthetic biology, particularly for engineering metabolic pathways that convert feedstocks into valuable products. Assessment of gene essentiality at a genome scale requires large and costly growth assays of knockout strains. Here we describe a strategy to predict the essentiality of metabolic genes using binary classification algorithms. The approach combines elements from genome-scale metabolic models, directed graphs, and machine learning into a predictive model that can be trained on small knockout data. We demonstrate the efficacy of this approach using the most complete metabolic model of Escherichia coli and various machine learning algorithms for binary classification.