My current research
My current research aims to understand how DNA is transcribed. Information about this process is encoded in the spatial distribution and motions of the molecules involved, which I study at the single molecule level through the use of super-resolution imaging and machine learning.
Localisation-based single molecule tracking techniques are central to my work. Individual target molecules (proteins and genes) are labelled with fluorescent probes which are excited one molecule at a time. By fitting a mathematical function to the observed fluorescent spots, each molecule can be localised with a resolution of approximately 20 nm, beyond the diffraction limit of light, and tracked as it performs its function in living cells. This reveals molecular behaviours, enabling fundamentally important biological questions to be tested. I have subsequently built further on these techniques using computational, machine learning, and optical approaches to make observations allowing the study of sequences of events from the point of view of a single molecule.
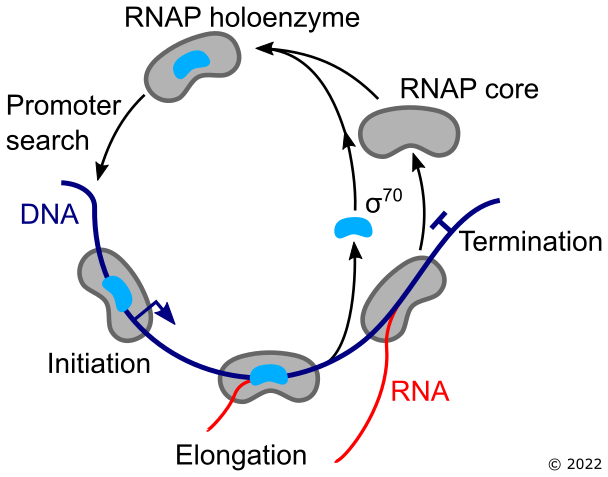
Central to transcription is a protein machine known as RNA polymerase (RNAP), which transcribes genes encoded in DNA into messenger RNA, the instructions for making new proteins. This process exists in all known forms of life. The targeting of this machine to specific groups of genes is performed by transcription factors, such as the bacterial sigma factor σ⁷⁰. Interactions between these proteins underpin gene expression. I am currently working to understand this system, and test ideas around how these components function in the real world.
Custom Instrumentation
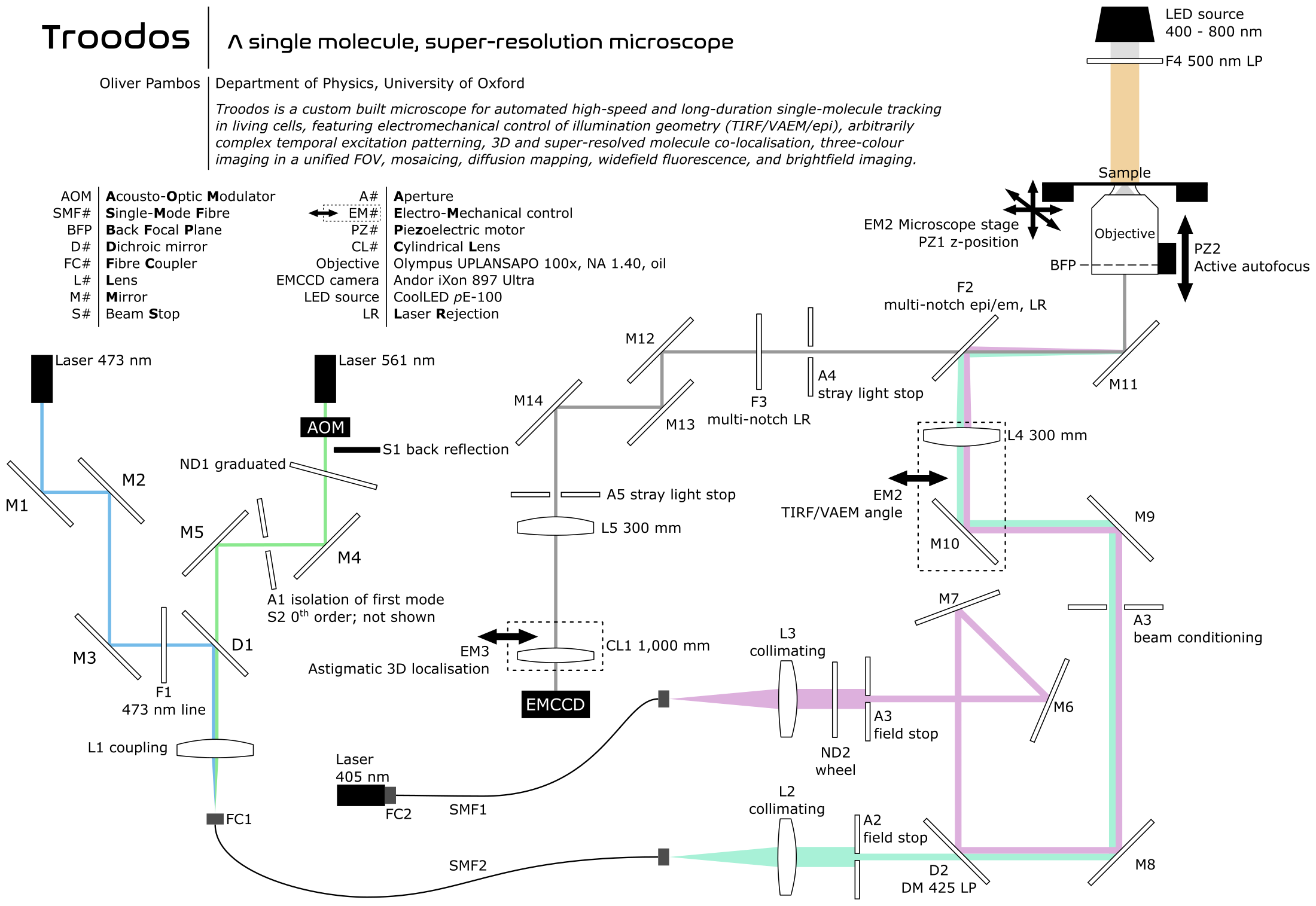
To enable this work, I designed and constructed a custom-built super-resolution microscope (Troodos) for high-speed and long-duration single-molecule tracking in living cells. The system provides precise control over illumination geometry and excitation timing, allowing individual molecules to be localised and tracked with extremely high precision over temporal scales from milliseconds to tens of minutes.
Originally developed for my own research, this microscope has since become a core experimental platform for a wide range of experiments across multiple laboratories within Oxford's Department of Physics, Biochemistry, and the Kavli Institute for Nanoscience Discovery, underpinning publications spanning single molecule protein and gene tracking, liquid–liquid phase separation, plasmid localisation, phage invasion, horizontal gene transfer, and single-molecule tracking in eukaryotic systems.