Research interest
I have a background in biology and I am largely interested in the evolutionary process.
With Ard Louis, I am working on furthering understanding of genotype-phenotype maps, i.e. the relation between genetic information and the phenotypes they produce (functions, structures and behaviours). In my own projects, I currently work with molecular genotype-phenotype maps such as the well-studied RNA folding landscape. More excitingly, I am exploring the protein folding landscape, which can only be studied now that tools for fast protein structure prediction have become available (AlphaFold, ESMFold).
When studying the (evolutionary) properties of genotype-phenotype maps, one exciting theme is the evolution of complexity (at all scales of organization in biology). Prior work has shown a strong bias in evolution towards simple phenotypes. The role selection plays in this simplicity bias is still debated, and similarly the multi-levelness of biology (molecules, cells, organisms) and various other details of biological reality have not been integrated into this picture. Such factors may help explaining why complexity does evolve at times, even if rarely: it took roughly 2 billion years for simple cells (prokaryotes) to evolve into complex cells (eukaryotes).
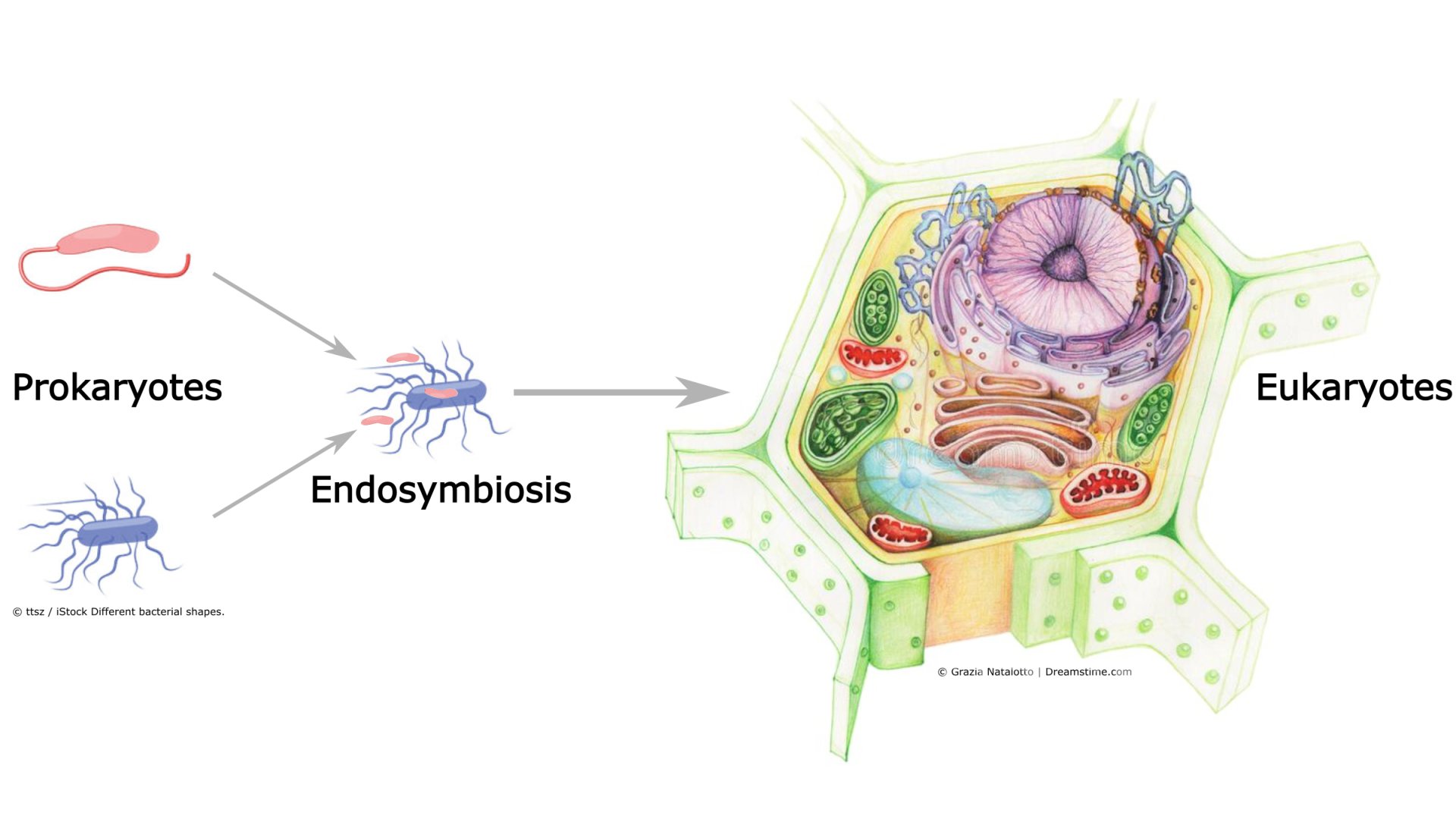
Eukaryotes (complex cells) originate from the merger between two distinct prokaryotes some 1-2 billion years ago.

